Developing antivirals in a rational way: “blunting” the SARS-CoV-2 scissors
The ongoing COVID-19 pandemic has forced the scientific community to devote itself to the research and development of various strategies to fight SARS-CoV-2 infection. The only informed way to identify new antivirals is through the knowledge of the three-dimensional structure, literally the shape, of the virus proteins that are responsible for cell infection and virus replication. Once the structure of proteins is known, it is possible for chemists to synthesize compounds that block its function, and consequently the virus life cycle.
We talk about it in this cycle of 5 episodes with the help of Dr. Massimo Degano, Group leader of the Biocrystallography Unit of the IRCCS San Raffaele Hospital and lecturer at our University (he teaches Chemistry at the Degree Course in Dentistry and Dental Implantology and at the International MD Program, Biochemistry at the Degree Course in Medicine, and Structural Biology at the Degree Course in Biotechnology Research in Medicine).
How many times have we wished in recent months that specific antivirals against SARS-CoV-2 could be developed soon? Many times, obviously – and rightly so. But how can we make this wish come true? What strategies can we use, taking advantage of the information that derives from the isolation and sequencing of the virus’ genetic material?
First, we need to identify a molecular target we can aim our weapons to. And here it is of paramount importance to have previous knowledge of Coronavirus biology. Incidentally, up to twenty years ago Coronaviruses were considered not very dangerous, and those who studied them seemed to be interested in a problem with limited applicability for human health. It is thanks to these studies guided by the curiosity of many scientists, basic research, that today we have a compass to guide our movements in the battle against the virus.
When Coronaviruses infect cells, they release their genetic material (a large RNA molecule) and this is used by our cellular machinery – literally hijacked – to synthesize the proteins of the virus. The virus itself is organized in such way that some of its proteins are synthesized joined together, like pearls on a necklace. To form new viral particles, proteins need to detach from each other. And here molecular scissors intervene, the viral proteases which, by cutting the necklace in specific points, release the proteins to allow each one to perform its function.
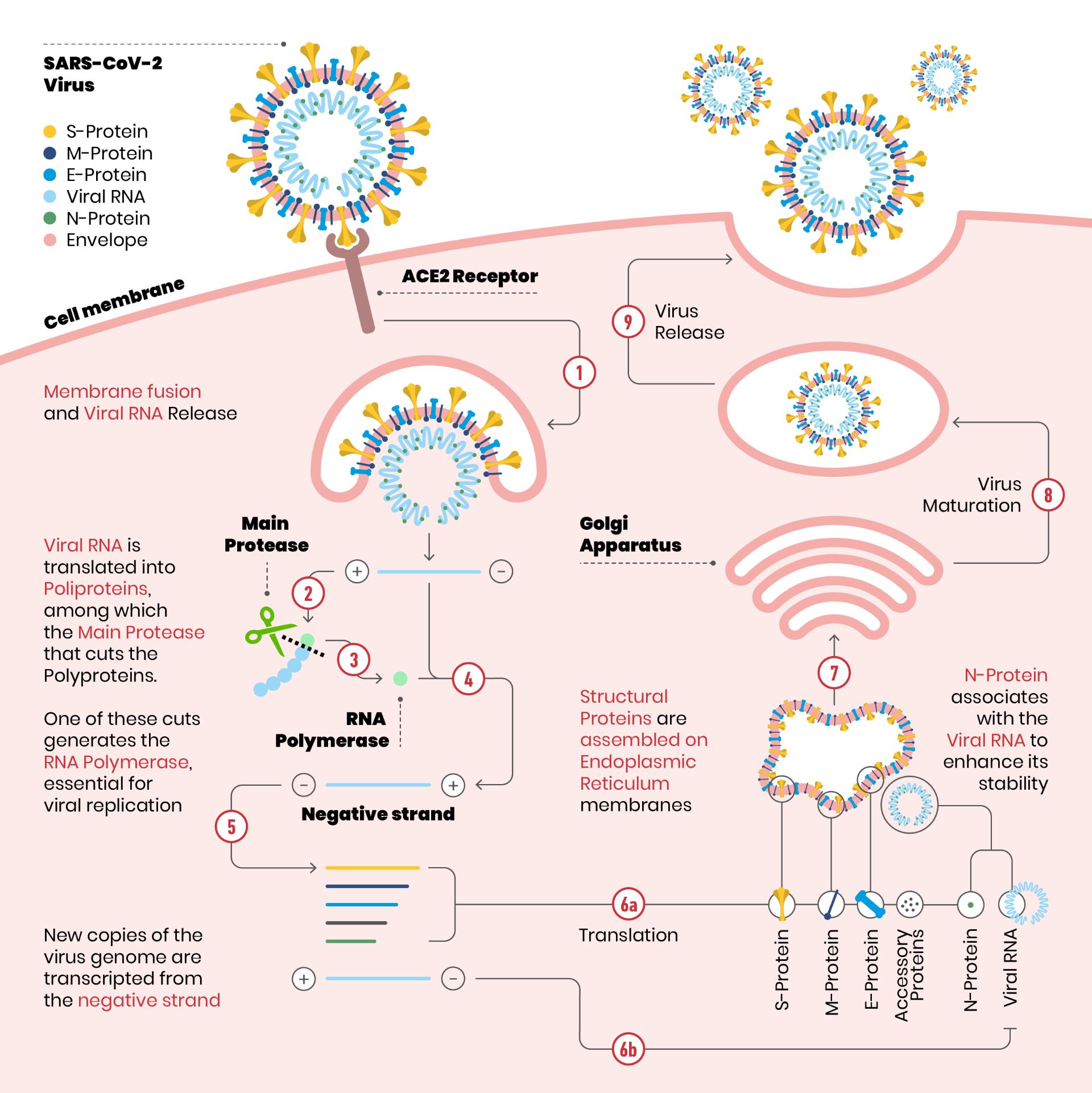
Scheme illustrating SARS-CoV-2 replication cycle
The main protease of SARS-CoV-2 is called Mpro; without its action, the virus cannot replicate and infect new cells! Therefore, a molecule that “blunts” these viral scissors would be a compound with potential antiviral action. These compounds are called inhibitors because they block the natural activity of the protease. Mpro is an even more attractive target because of its uniqueness, i.e. the fact that in the human body there are no proteins with a similar structure and function, so a compound that inhibits the action of the viral protease would be less likely to have serious side effects on the patient.
How do we engineer a molecule that inhibits Mpro? If you are following us in this series, by now you know the answer: nothing better than visualizing the structure of the protein, seeing which portion of it constitutes the “blade” of the scissors and what shape it has in order to build something that prevents it from carrying out its molecular cuts. What we want is to obtain a compound that has high affinity towards the protein, i.e. that is able to inhibit Mpro, even if present in small quantities. Furthermore, it is important that it has high specificity, i.e. avoiding its distraction from the task by other proteins in the cells.
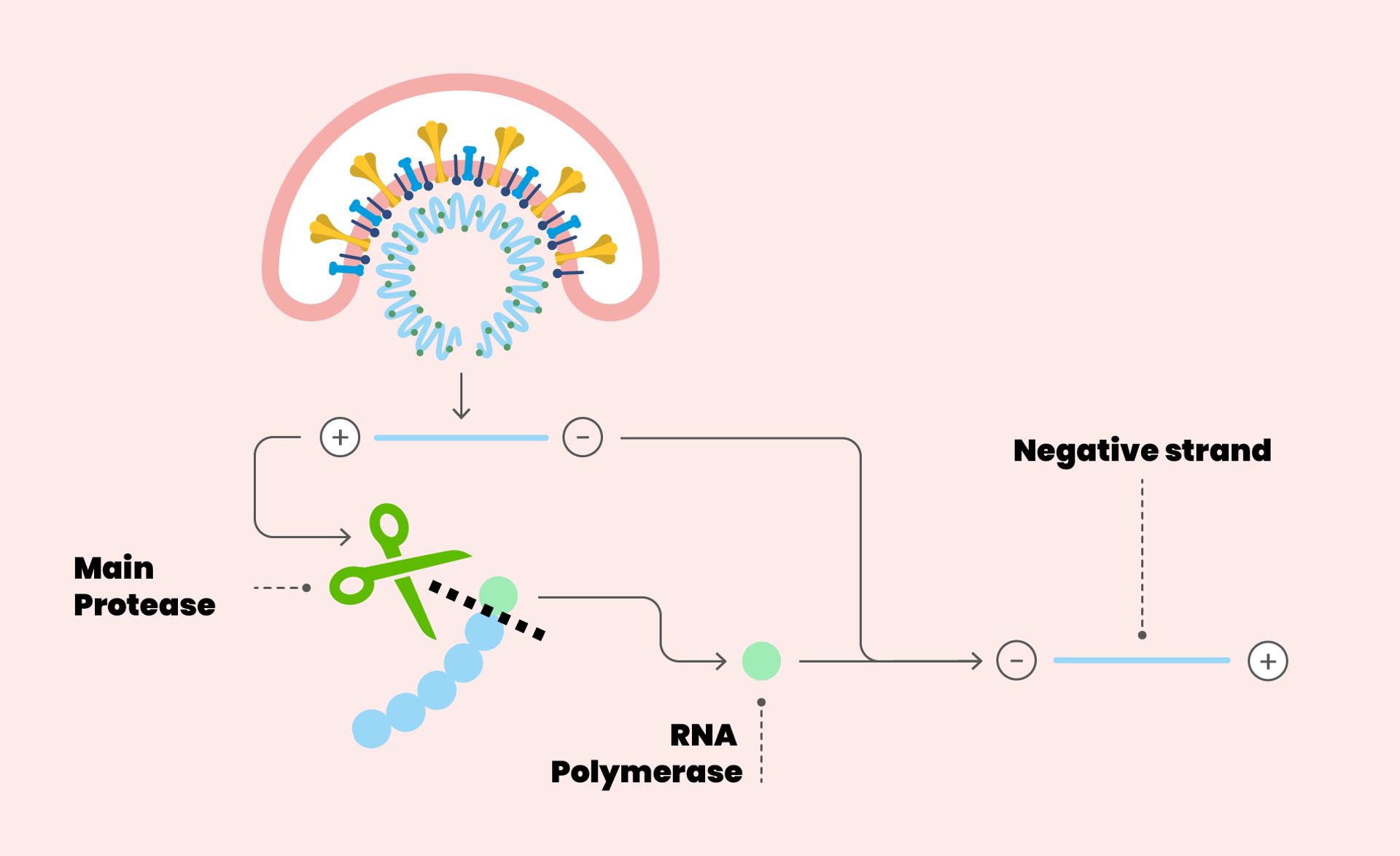
Detail of the SARS-CoV-2 replication cycle. The envelope of the virus merges with the cell membrane. Once the RNA is released into the cytoplasm, it is translated into polyproteins, including the main protease (the scissors in figure). The protease makes cuts on the polyproteins: one of these cuts originates the enzyme RNA polymerase, essential for the replication of the virus.
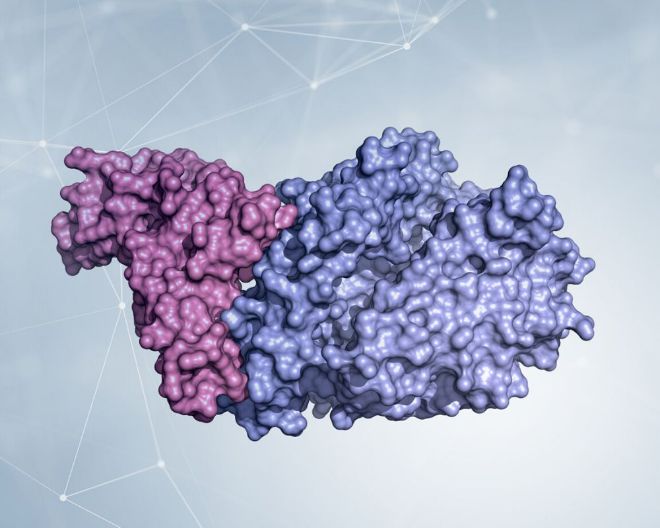
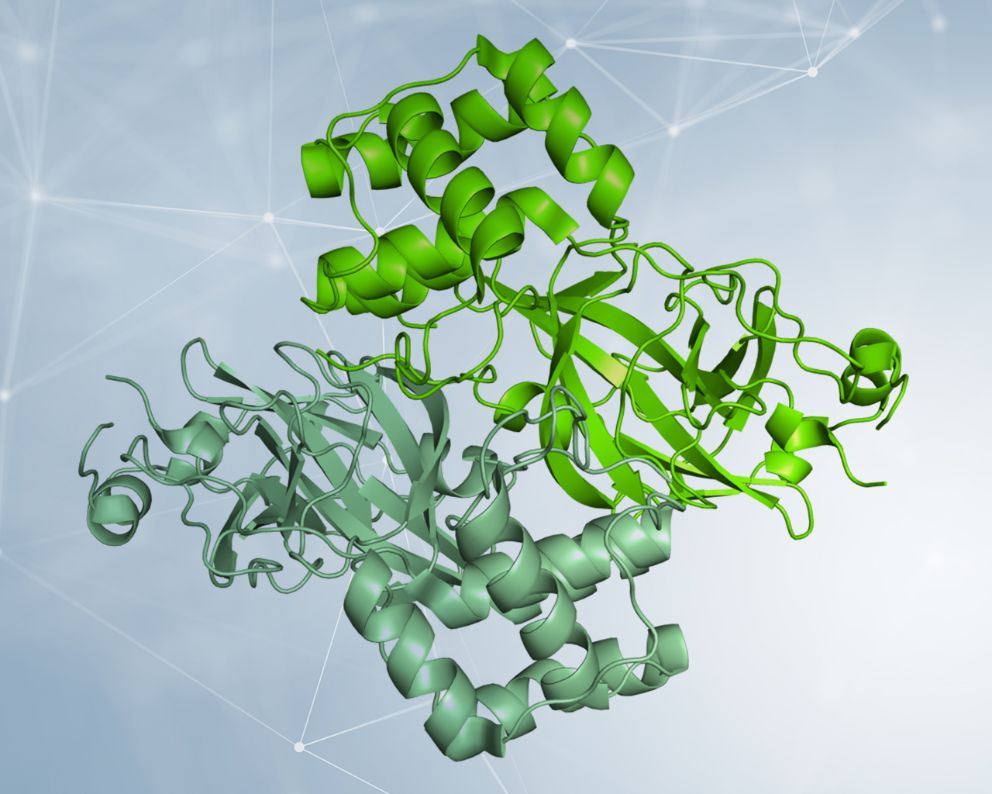
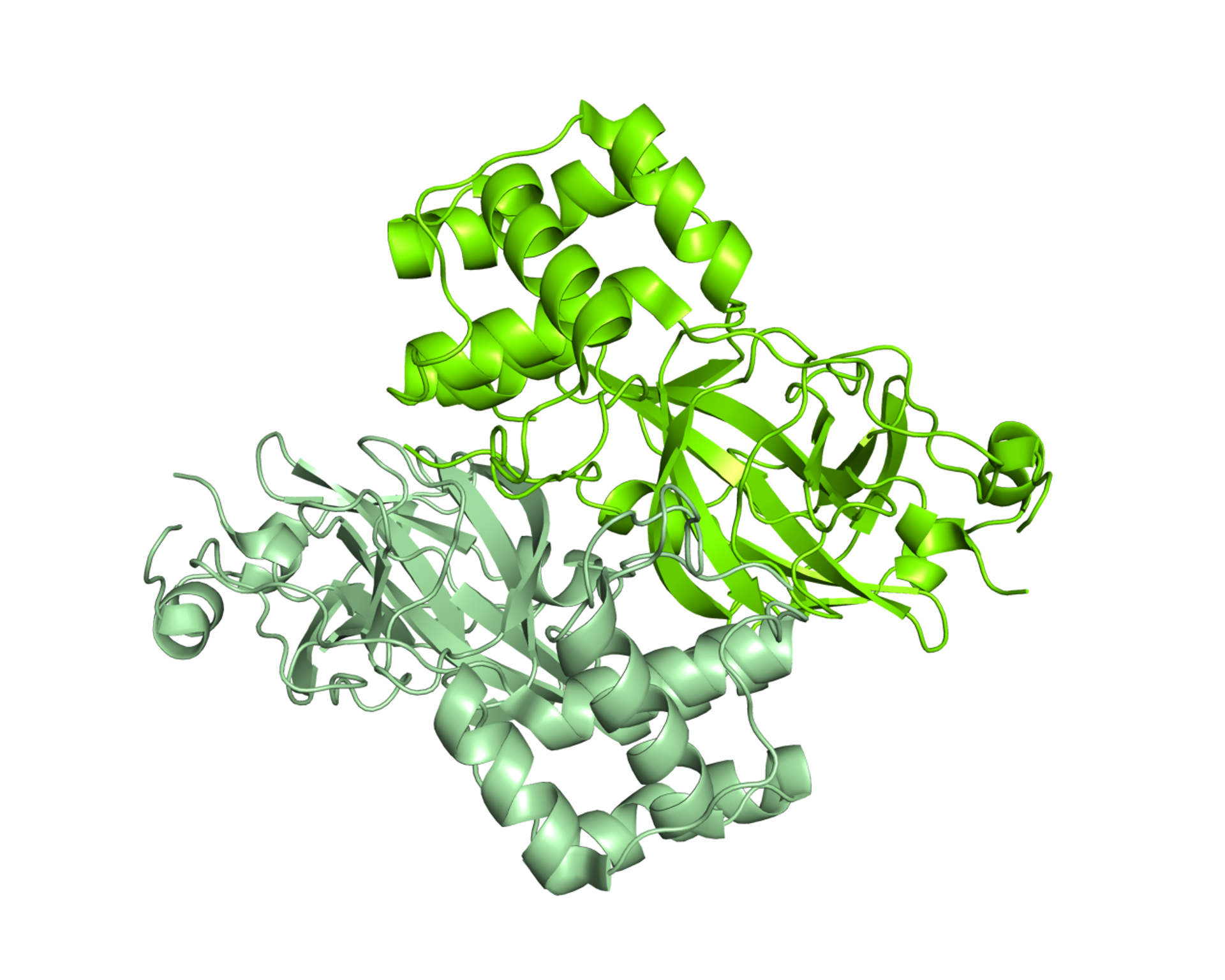
Already in February the research groups of prof. Rao and Hilgenfeld determined the first Mpro structures from SARS-CoV-2 [1] both in its form of “open scissor” and linked to various inhibitors, one of which is highly specific and with high potential to represent an antiviral future [2]. The Mpro protease is a dimer, i.e., in order to function it must be made up of two equal chains associated with each other, and it possesses a pocket at the surface where the viral polyprotein is cut. The characteristics of this pocket (shape, size) must be used to engineer an optimal inhibitor.
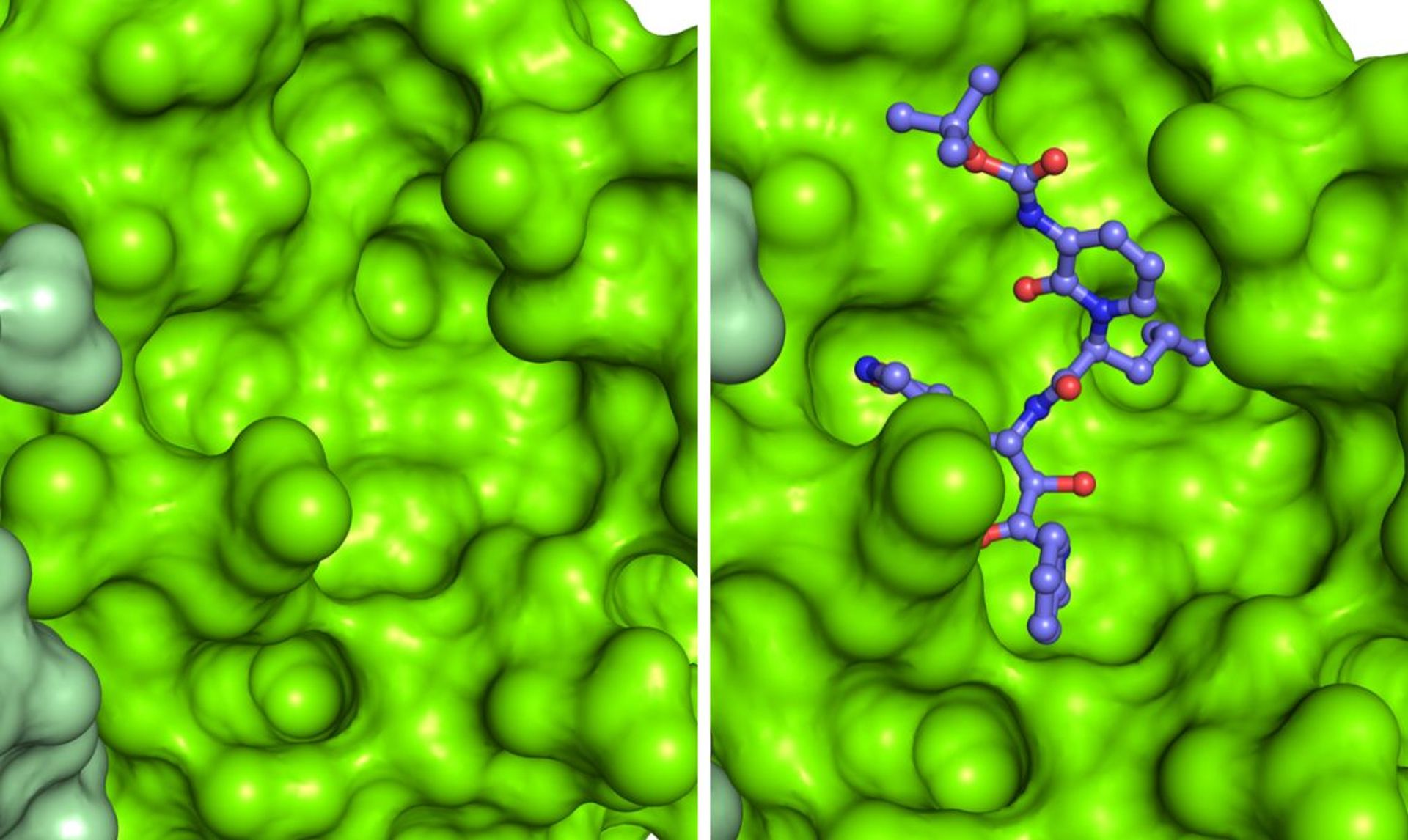
Detail of the dimeric structure of Mpro from SARS-CoV-2, represented as a molecular surface, with the two chains colored in two shades of green. The analysis of the structural characteristics of the protein made it possible to identify a surface pocket in which the viral polyprotein is cut, and to engineer inhibitors. An example is represented by a compound complementary in form and chemical characteristics to the surface pocket and which inhibits the Mpro activity. The structure used to make the images is deposited with the code 6Y2F in the Protein Data Bank (http://www.rcsb.org/pdb). Courtesy of Dr. Degano.
At the present time, 152 Mpro structures have been determined linked to different fragments of molecules that give indications on how to improve the affinity and specificity of the inhibitors. In this case the technique used is the X-ray crystallography: since 1945 it has continued to allow us to visualize biological molecules with a detail down to the level of the single atom. In the next episode we will see what crystallography is and how it works!
Read the other episodes of the series "Structural biology for SARS-CoV-2"
First episode: "The Spike protein"
Second episode: "Cryo-electron microscopy: small electrons to visualize large molecules"
Fourth episode: "Structural biology for SARS-CoV-2: X-ray crystallography"
Fifth episode: "RNA polymerase, the SARS-CoV-2 “sloppy photocopier”"
References
[1] Jin et al. “Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors” (2020) Nature 582, 289-293. DOI: https://doi.org/10.1038/s41586-020-2223-y
[2] Zhang et al. “Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors” (2020) Science 368, 409-412. DOI: 10.1126/science.abb3405
You might be interested in
Rapid and portable tests for COVID-19: UniSR partner of the CORONADX project
Structural biology for SARS-CoV-2: X-ray crystallography
Cryo-electron microscopy: small electrons to visualize large molecules